1st column is the file name. The "
Go to PubPeer" link will work perfectly if the file name is a DOI, a PMID or an arXiv ID.
2nd column gives the PMID of the closest neighbor (according to inter-textual distance
[1]) in the reference set (see
[2] for description of this set). First line of the cell can be one of the following.
"
Far" means the tested paper is far enough (dist. greater than 0.5) from its nearest neighbor in the reference set (thresholds description in
[2]).
"
Close" means there may be something to look for (dist. between 0.44 and 0.5).
"
Very Close" means that the tested paper and the one in the reference are very close (dist. lower than 0.44). They may share portions of text.
3rd column lists the gene identifiers found in the tested text. The numeric value in brackets is the number of occurrences of the identifier that was found within the text.
4th column lists possible contaminated cell lines
[3].
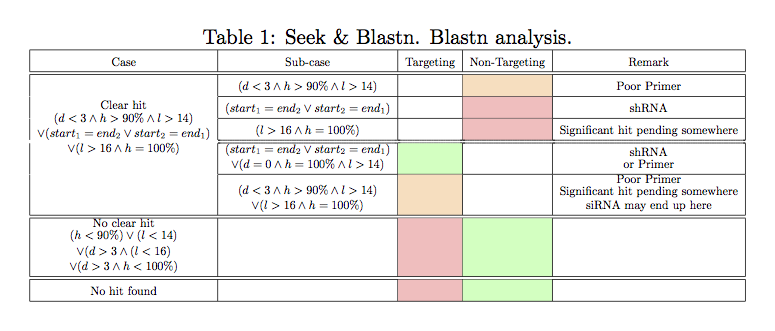
5th column gives blastn
[4] resluts as follows :
(gene name) Nucl. Seq. (Text Claims : gene (e-value / n-n / p) ... )(gene name) this is potentially the gene that the text is talking about for this sequence (unknown error rate)
Nucl. Seq. The blasted Nucleotide Sequence with an hyperlink to a Google query for this sequence
Text is a link to the place (in the pdf file) where the sequence possibly appears.
Claims is the claimed status can be: "
Claims targeting", "
claims non-targeting" or "
Undetected claim".
gene (evalue / n1-n2 / p) ... is a summary of the blastn result (
Nucl. Seq. is the query sequence for the blastn query):
The hyperlink gives access to the detailed balstn results.
"
!!" and red color means that the result found by seek & blastn is in contradiction with the claimed status. Green color means no contradiction. Orange means questionable.
"
No hits found" means no hits found, "
No clear target" means that no significant target has been found (see
details).
gene, is a gene for which a significant hit has been found (see
details).
e-value, is the e-value (see blastn doc for
e-value signification) of the hit. For each gene, only the hit with the smallest e-value is given.
n1-n2,
n1 is the place where the hit ends in the query seq. and
n2 is the size (nucleotide length) of the query seq (
Nucl. Seq.).
p is the % of identities for this hit.